Measuring cells isolated from blood or tissues provides an enormous amount of information on a per cell basis. However, the removal of cells from these tissues involves the destruction of the tissue architecture and microenvironment in which these cells originated. The activity of immune and haematopoietic cells are largely driven by the microenvironment in which they find themselves, both in haematopoietic/immune tissues (e.g. bone marrow, lymph node) and inflammatory sites (lung, CNS).

As such, the use of HD imaging technologies is crucial to mapping inflammatory responses over time. Novel imaging systems, such as Imaging Mass Cytometry (IMC), can measure up to 40 antibodies or transcripts on tissue sections in an automated fashion at approximately 0.75 mm^2 per hour at 1 um resolution.
Spatial analysis with Spectre & SpectreMAP
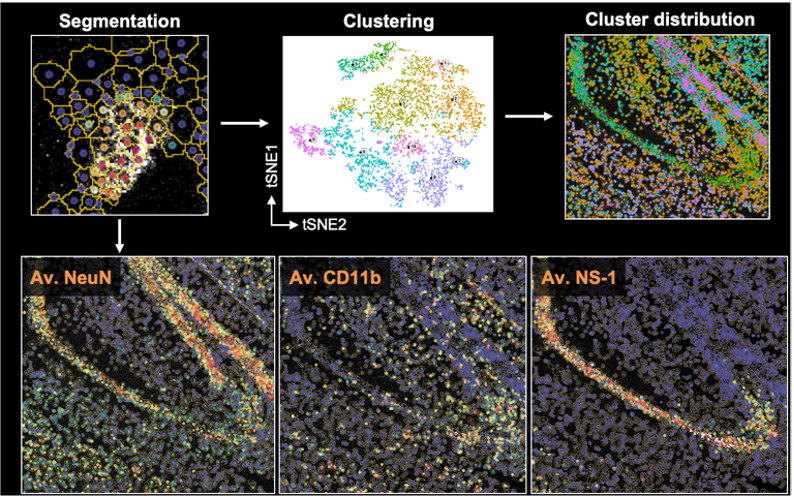
Along with flow, spectral, or mass cytometry data, we have extended Spectre to facilitate spatial analysis of Imaging Mass Cytometry (IMC) data. To do this we developed an extension of Spectre, termed ‘SpectreMAP’, to import, manage, and visualise TIFF files using RStudio. Once cell segmentation has been performed (using our protocols, or those developed by others), the marker expression data for each cell across multiple images can be calculated and incorporated into a single data.table. Clustering, gating, or image-based classification can then label cellular populations. Subsequently, the relationship between cell types and their spatial arrangement and environments can be calculated using our spatial analysis functions. For an overview of the spatial analysis components, please see this page. Our ongoing work is focused on the development of better data integration approaches and spatial analysis tools.
Cell segmentation
A significant challenge in the analysis of spatial data is the segmentation of the image into individual cells. Because of the substantial overlap between neighbouring cells in imaging data, this process is not trivial. We are developing adaptable approaches to cell segmentation in cell-dense tissues, through the use of Ilastik and other tools, largely build on the use of classifiers to predict cell boundaries based on expression data from multiple markers simultaenously.